An IGNITE-funded research article has been published in the Nature Communications journal and looks into the animal genomic evolution with the chromosomal-level assembly of the freshwater sponge Ephydatia Muelleri. The genomes of non-bilaterian metazoans are key to understanding the molecular basis of early animal evolution. However, a full comprehension of how animal-specific traits, such as nervous systems arose, is hindered by the scarcity and fragmented nature of genomes from key taxa, such as Porifera.
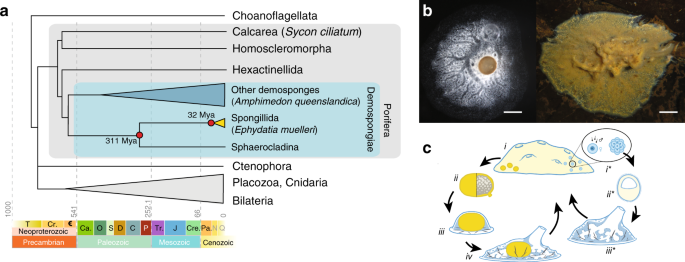
The study confirms that Ephydatia muelleri is a freshwater sponge found across the northern hemisphere. It has a global distribution and century-long history of study both in situ and in the laboratory, and as such E. muelleri raises numerous questions about adaptation and the evolution of animal characters.
The team of researchers, examines throughout the study the chromosome-level assembly of the 326 million base pair (megabases, Mb) Ephydatia muelleri genome. The highly contiguous assembly of this sponge genome is an exceptionally rich resource that reveals metazoan typical regulatory elements, macrosynteny shared with other non-bilaterians and chordates, and allows analysis of structural chromatin variation across animals. The high gene count of sponges compared to other animals is shown to be a feature of gene duplication. These analyses, together with the evaluation of RNA expression in development and host–microbe relationships across the range of this species provide key data for understanding the genomic biology of sponges and the early evolution of animals.
The paper analyses reveal a metazoan-typical genome architecture, with highly shared synteny across Metazoa, and suggest that adaptation to the extreme temperatures and conditions found in freshwater often involves gene duplication. The pancontinental distribution and ready laboratory culture of E. muelleri make this a highly practical model system which, with RNAseq, DNA methylation and bacterial amplicon data spanning its development and range, allows exploration of genomic changes both within sponges and in early animal evolution.
You can read the full paper here.